Research
Our multidisciplinary research approach combines concepts from physics, population genetics and cancer biology with innovative genetic tools and sophisticated cellular model systems to study evolution in dense cellular populations. Using complementary microbial and cancer cell assays, we investigate (i) how collective multicellular dynamics impact genetic diversity and drug resistance evolution, (ii) how collective cell motion in 3D affect evolution and treatment response, and (iii) how the mechanical properties of cells and their interaction with the biophysical microenvironment reshape evolutionary trajectories in cancer tumoroids.
Our goal is to establish a new framework describing cellular evolution as an emergent phenomenon in active granular matter, providing a springboard for innovative evolution-based treatment strategies for improved cancer therapy.
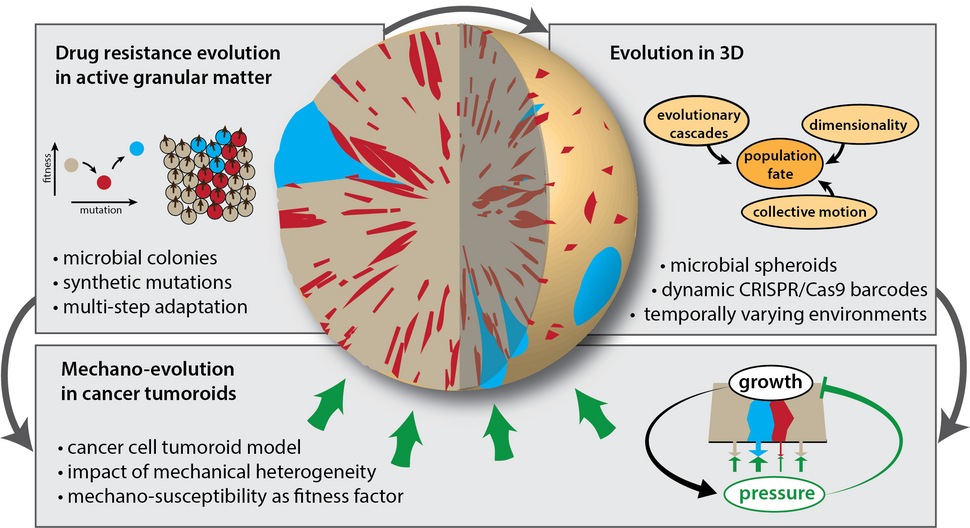
Multi-step drug resistance evolution in active granular matter
Dense, growing populations of cells can be described as a form of active granular matter. A hallmark of these systems is that the relative motion of individual cells can become correlated over long length scales. How such correlations shape the fundamental mechanisms of evolution, such as genetic drift and natural selection, remains unclear. We use a genetically tailored yeast-based model systems, featuring tunable “synthetic mutations”, to study how density-induced multicellular dynamics restructure genetic population diversity, facilitate the evolutionary rescue of less-fit drug resistant clones and allow populations to adapt across substantial fitness barriers.
Collective cell dynamics, evolution and therapy response in 3D populations
Many dense cellular populations, including solid tumors, grow 3-dimensionally. This is in stark contrast to most established experimental evolution assays for range expansions, typically based on 2-dimensional, surface-bound microbial colonies. While theory and computational models of evolution suggest a marked difference between 2D and 3D range expansions, we currently lack the means for a paralleling empirical investigation. We recently established matrix embedded microbial spheroids as a new tool for experimental evolution. Leveraging the versatility of this technique in conjunction with our synthetic mutations toolbox, we study how collective cell dynamics change the genotypic heterogeneity and ensuing treatment resilience of three-dimensionally expanding populations. In addition, this assay serves as an ideal platform to design and test new evolution-based therapy strategies to mitigate the consequences of drug resistance evolution.
Mechano-evolution and adaptive therapy in cancer tumoroids
Changes in mechanical cell properties and an altered response to physical stresses are both a hallmark of cancer cells, with many tumors exhibiting a vast of heterogeneity in both features. Yet, little is know how these mechanical aspects of tumor expansion impact key evolutionary processes, such as natural selection and genetic drift, and critical biomedical consequences, including the competitiveness of metastatic precursor cells inside the primary tumor. We designed a new mammalian-cell tumoroid model system, tailored for experimental evolution, to investigate the fundamental dynamics of mechano-evolution in a biophysical microenvironment. A specific focus of this project is the impact of mechanical population heterogeneity and the fitness effects of altered mechano-sensitivity on cancer evolution. The gained insights serve as the starting point for a reinforcement-learning approach towards improved adaptive cancer therapy.
Contact
Guck Division
Kayser Junior Research Group
MPI for the Science of Light
Staudtstr. 2
D-91058 Erlangen, Germany





