Real-Time Deformability Cytometry

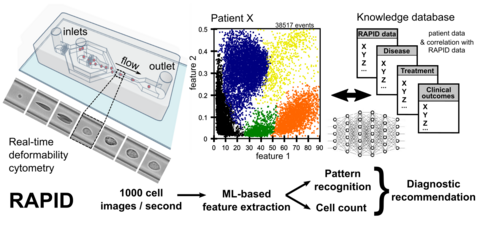
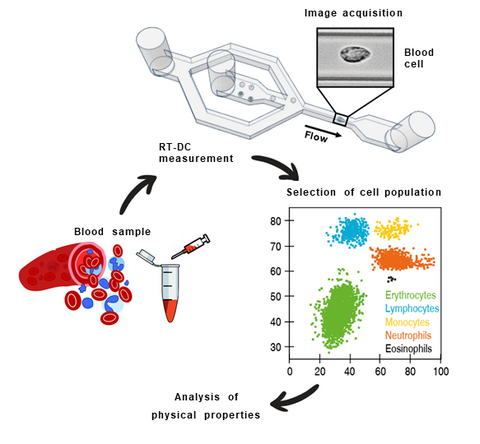
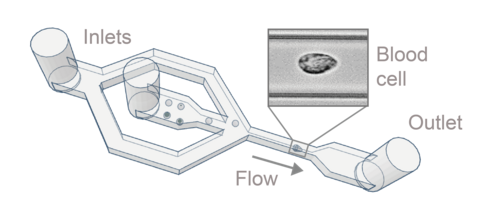
Real-time deformability cytometry (RT-DC) is a novel high-throughput method for the mechanical characterization of single cells that has recently been developed in our lab [3, 4, 5]. Based on the hydrodynamic deformation of cells translocating through a microfluidic channel in a contact-free manner, RT-DC is able to analyze more than 100 cells per second in real-time.
The main working principle of RT-DC is shown in the figure. Within a few milliseconds upon entry of a cell into the channel, the cell shape reaches a steady state. For each cell, several parameters can be recorded in real-time which can be visualized and gated in a post-processing step using our in-house software ShapeOut. Analytical and numerical models developed in our lab also permit the derivation of material properties such as the Young’s modulus [1, 2]. The technique is now being applied in more than 50 collaborations [6, 7, 8].
Furthermore, we have recently developed an RT-DC setup with fluorescence detection (RT-FDC). Now it is possible to not only detect the mechanical phenotype of each individual cell but also to simultaneously gather its fluorescence intensity in up to three channels in a manner similar to a conventional flow cytometer [9]. This direct correlation of mechanical with fluorescence data based on dyes, fluorescent reporter proteins or surface markers will lead to a more comprehensive validation of cell mechanics as a label-free marker.
RT-DC and RT-FDC are available as commercial products from the spin-off company ZELLMECHANIK DRESDEN GmbH.
For the sake of science communication, the following media, all related to deformability cytometry, are dedicated to the public domain (CC0):
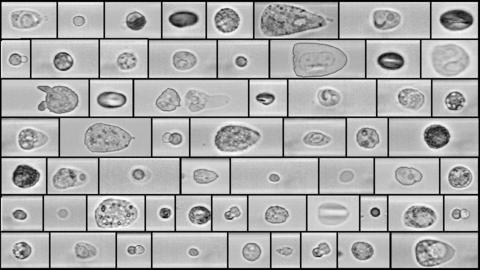
[1] A. Mietke, O. Otto, S. Girardo, P. Rosendahl, A. Taubenberger, S. Golfier, E. Ulbricht, S. Aland, J. Guck, and E. Fischer-Friedrich, “Extracting cell stiffness from real-time deformability cytometry: theory and experiment,” Biophysical Journal, vol. 109, iss. 10, p. 2023–2036, 2015.
[2] M. Mokbel, D. Mokbel, A. Mietke, N. Träber, S. Girardo, O. Otto, J. Guck, and S. Aland, “Numerical simulation of real-time deformability cytometry to extract cell mechanical properties,” Acs biomaterials science & engineering, 2017.
[3] O. Otto, P. Rosendahl, A. Mietke, S. Golfier, C. Herold, D. Klaue, S. Girardo, S. Pagliara, A. Ekpenyong, A. Jacobi, M. Wobus, N. Töpfner, U. F. Keyser, J. Mansfeld, E. Fischer-Friedrich, and J. Guck, “Real-time deformability cytometry: on-the-fly cell mechanical phenotyping,” Nature Methods, vol. 12, iss. 3, p. 199–202, 2015.
[4] M. Herbig, M. Kräter, K. Plak, P. Müller, J. Guck, and O. Otto, “Real-time deformability cytometry: label-free functional characterization of cells,” in Flow cytometry protocols, Springer New York, 2017, p. 347–369.
[5] M. Urbanska, P. Rosendahl, M. Kräter, and J. Guck, “High-throughput single-cell mechanical phenotyping with real-time deformability cytometry,” in Methods in cell biology, Elsevier, 2018, p. 175–198.
[6] N. Toepfner, C. Herold, O. Otto, P. Rosendahl, A. Jacobi, M. Kräter, J. Stächele, L. Menschner, M. Herbig, L. Ciuffreda, L. Ranford-Cartwright, M. Grzybek, Ü. Coskun, E. Reithuber, G. Garriss, P. Mellroth, B. Henriques-Normark, N. Tregay, M. Suttorp, M. Bornhäuser, E. R. Chilvers, R. Berner, and J. Guck, “Detection of human disease conditions by single-cell morpho-rheological phenotyping of blood,” eLife, vol. 7, 2018.
[7] M. Kräter, J. Sapudom, N. Bilz, T. Pompe, J. Guck, and C. Claus, “Alterations in cell mechanics by actin cytoskeletal changes correlate with strain-specific rubella virus phenotypes for cell migration and induction of apoptosis,” Cells, vol. 7, iss. 9, p. 136, 2018.
[8] M. Urbanska, M. Winzi, K. Neumann, S. Abuhattum, P. Rosendahl, P. Müller, A. Taubenberger, K. Anastassiadis, and J. Guck, “Single-cell mechanical phenotype is an intrinsic marker of reprogramming and differentiation along the mouse neural lineage,” Development, vol. 144, iss. 23, p. 4313–4321, 2017.
[9] P. Rosendahl, K. Plak, A. Jacobi, M. Kraeter, N. Toepfner, O. Otto, C. Herold, M. Winzi, M. Herbig, Y. Ge, S. Girardo, K. Wagner, B. Baum, and J. Guck, “Real-time fluorescence and deformability cytometry,” Nature methods, vol. 15, iss. 5, p. 355–358, 2018.